Introduction

Species
distributional patterns are neither stable nor random, and follow
geographic patterns that reflect both recent ecological conditions
and processes and phylo-geographic history (Avise 2000). At present
time, species that originated in the past as small disjunct
populations seem to have rearranged their current distributions
showing a sympatric or parapatric assemblage in areas of high current
capacity (Fjeldså 1995). Thus, present distribution of the
Andean biota reflects today's biotic and abiotic interactions
of the species and the ecosystems in which they occur. As a result,
species distribution and accumulation in such areas seems to be
related to a sharp replacement of different habitat types within
short distances (Fjeldså et al. 1999; García-Moreno
et al. 1999).
Because distribution
patterns are not homogeneous, it is more likely to argue the presence
of specific environmental conditions within the carrying capacity
areas that favor species accumulation. According to Boone et al.
(2000) the spatial heterogeneity observed at present in biotic
communities is an expression of the biotic and abiotic factors that
shape the n-dimensional space of the specie's niche (Hutchinson
1957). Subsequently, the spatial distribution of Andean centered taxa
could be explained by the environmental gradients present on the
Andean chain.
To understand
patterns of species diversity, it is necessary to evaluate the
temporal and spatial processes that have shaped their current
distribution and the ones that sustain them at present time (Heindl &
Schuchmann 1998; Huston 2002). Thus,
generating detailed and accurate distributional information for such
species becomes a critical step in such understanding process
(Peterson et al. 2002).
Museum and herbarium
specimens constitute the primary documentation of a species´
true presence records. However, information of species´ true
absence is almost nonexistent. These drawbacks reflect on the
production of species geographic ranges represented by simple
collection point dots plotted over country base maps. In the best
scenario species geographic ranges are presented as shaded outline
maps that connects know collection localities (Anderson &
Martínez-Meyer 2004).
A potential
non-subjective approach to maximize the utility of available
distribution data for such areas is to develop species-distribution
models (SDMs). Recent advances employing geographic information
systems (GIS) technology allow correlative modeling of species'
potential macro-distributions (Anderson et al. 2002a). SDMs
are based on the assumption the relationship between a given pattern
of interest (e.g. species abundance or presence / absence) and a set
of factors assumed to control it, exists and can be quantified
(Guisan & Zimmermann 2000; Huston 2002). Correlative approaches
are inspired by the Hutchinson's fundamental niche concept
(1957) and are capable of predicting hypothetical distributions based
on ecological and environmental requirements of species,
extrapolating from known sample points into unknown areas (Anderson
et al. 2003; Raxworthy et al. 2003; Anderson &
Martínez-Meyer 2004).
Most
correlative models are based on the assumption that the observed
pattern of species occurrence is in equilibrium with the environment,
and that this relationship is static in time (Guisan 2002). However,
species distributions respond to time complex processes (i.e.
exclusive competition) that prevent species from fully occupying
their potential ranges (Williams et al. 2002). In spite of
these limitations, correlative models are useful to model species
distributions for large geographic areas, especially when detailed
information about the natural history of species is lacking (Peralvo
2004).
The primary
objective of this study was to predict the species geographic
distribution of nine Andean-centered genera using genetic algorithms
on the Eastern Mountain Range (Eastern Cordillera).
Materials and Methods

Study Area
The Northern Andes
forms a complex system of 14 sub-regions defined by their
characteristic set of natural communities that share a large majority
of their species, dynamics, and environmental conditions (Dinerstein
et al. 1995; WWF 2001). The Eastern Cordillera Real Montane
Forest and the Northern Andean Paramo are two of such areas and both
were chosen as our study area (Eastern Cordillera hereafter) due to
its high level of neo-endemic species diversity.
The Eastern
Cordillera has its southern limits in northern Peru, in an area where
the chain is bisected by the Huancabamba depression near the area
known as the Abra de Porculla at approximately 6ºS. This area
constitutes a strong barrier that divides biogeographically the Andes
in two different regions (Duellman & Wild 1993; Myers 2000). From
this point the Andes extent northeast through Ecuadorian territory
until it reaches the eastern drainage of the Macizo Colombiano
(Colombian massif) at 2ºN of latitude covering an area of
approximately 9,236 km2.
The limit on the
eastern versant of the Andes was set at 500 m of elevation to exclude
the lowland Amazonian biota following previous studies of Andean
plant's distribution (Jorgensen et al. 1999; Borchsenius
1997; Vargas et al. 2004). The western limit is rather
arbitrary and tracks an irregular contour line at 3000 meters alt.
following remnant distribution patterns of the upper montane forest
of the western versant of the Eastern Cordillera (Figure1).
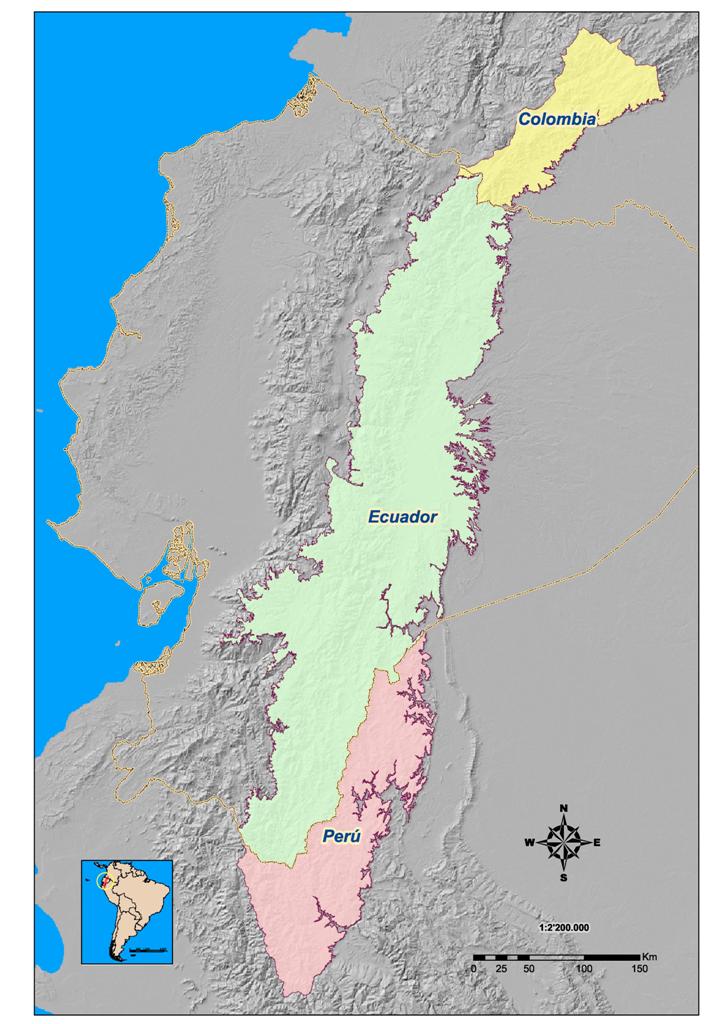
Figure 1. Location
of the Eastern Cordillera in South America.
Distribution modeling
The process for
distribution modeling relies on three main factors: (a) selection of
species; (b) collection of presence points from museum specimens and
herbaria records; and (c) generation of potential distribution maps
for the selected species using GARP.
a. Selection of
species
To understand the
significance of a biodiversity index across geography, one needs
context. Relationships between species and knowledge of lineages to
which they belong provide that context (Erwing 1991).
We chose a suit of
Andean-centered taxa (named Andean genera hereafter) that due to
their intrinsic characteristics can be used as good descriptors of
the diversity patterns of the Eastern Andean Ridge (Table 1). We
selected nine Andean genera as biodiversity subrogates considering
the following criteria: (1) their tempo and mode of radiation is
related with the Andean uplift and the climatic shifts of the
Pleistocene; (2) their overall distribution patterns present a high
level of replacement within the environmental gradients; (3) several
species of these genera present restricted distributions with a high
level of endemism.
Among vascular
plants we chose five genera that belong to the epiphytic or
hemi-epiphytic life form. According to Jorgensen & León-Yanez
(1999) more than a third of the plant species in the submontane and
montane forest rainforest of Ecuador belong to these types of life
form. Out of the total 2,110 native genera occurring in Ecuador the
five selected are among the 20 most diverse genera with more than 20%
of their listed species reported as endemics. This high level of
diversity within these groups is related to the elevation of the
Andes as well as to the climatic fluctuations during the Quaternary
(Gentry 1982; Croat 1991; Luteyn 2002).
Among vertebrate
groups we selected four Andean bird's genera (three
hummingbird's genera, and one antpitta genus). We used bird
species among others vertebrates because the available detailed
distributional data, a well defined taxonomic classification and
important literature dealing with their phylo-geographic patterns and
ecology (Bleiweiss 1998a, b; Fjeldså & Krabbe 1990; Ridgely
& Greenfield 2001; Rice 2005).
b. Collection of
presence points
We collected species
presence points from different datasets sources. Vascular plant
specimen data were obtained from the Missouri Botanical Garden's VAST
(Vascular Tropicos) nomenclatural database (MBG 2004), Herbarium
specimens from the Herbarium of the Instituto Nacional de Ciencias
(COL) in Colombia and the Catholic University Herbarium in Ecuador
(QCA). Bird species data were obtained from the following databases:
BirdLife International, Chicago Field Museum of Natural History
(FMNH), Academy of Natural Sciences of Philadelphia (ANSP) and the
Berkeley Museum of Natural History. Finally, additional data were
obtained in private data bases (Juan Fernando Freile for Antpittas;
Cal Dodson-Lorena Endara for orchid's records and James
Luteyn's database stored at the New-York botanical Garden site
(http://www.nybg.org) for
Ericaceae).
Out of these
databases, only those specimens whose collection localities were
designated with geographic coordinates were selected (topographic
maps 1:100.000 of the study area plus voucher level information were
used to geo-reference all specimens) and from these, only the
specimens identified at the species level by recognized experts of
the group were considered for the next steps and further analysis.
After obtaining the
presence records for each species of the selected Andean genera, the
number of species for developing the models within GARP was further
refined by choosing only those that had four or more geo-referenced
specimens (named unique points of occurrence hereafter) in the
biological databases.
The selection of a
minimum sample size of four localities was based on conciliation
between the data required to produce robust predictive models and the
fact that most of the species had very few unique collection
localities. However, other studies for not well explored areas have
used as few as five and four records respectively (Anderson et al.
2002b; Anderson & Martínez-Meyer 2004).
c. Species
Distribution Models (SDMs)
Generation of
accurate and reliable distribution maps is based on the quality of
the species occurrence points and on the selection and generation of
a good set of predictive variables. According to previous studies
(Leimbeck et al. 2004; Vargas et al. 2004; Luteyn 2002;
Borchsenius 1997; Schuchmann et al. 2001; Heindl &
Schuchmann 1998; Young et al. 2002; Jorgensen & León-Yanez
1999; Peralvo 2004; Boone & Krohn 2000) climatic and topographic
variables (environmental variable hereafter) are the most important
indirect predictors of present species assemblage within the Andes.
We selected a set of
eleven environmental variables for plants and nine for birds
considering the selected set as good indirect and direct descriptors
of the patterns of abundance and distribution of the Andean biota
(Table 2). The rationale of the selected set of environmental
predictors is a balance between variables tested to influence species
macro-distribution and the available information of environmental
factors that can be translated into a continuous raster format within
a regional geographic extent.
Each variable was
created in raster format with 500 m resolution using the GIS software
ArcGIS version 8.1 (Environmental Systems Research Institute,
Redlands, California). The 500 m resolution was chosen as an
intermediate point between the high resolution of the Digital
Elevation Model (DEM) used to generate the elevation variable (90 m,
see below) and the coarse resolution of the climatic variables. The
elevation variable was generated using the DEM produced by the Radar
Shuttle Topographic Mission (SRTM) at 90 m of resolution for the land
areas of the world. The DEM for the study area was downloaded using
the USGS Seamless Data Distribution System interface
(http://seamless.usgs.gov), and then it was re-sampled to a 500 m
resolution.
To refine the
resolution of the crude climatic variables an interpolation method
was applied to smooth the original resolution of 1 km2 to a
resolution of 500 m using an Ordinary Kriging (Kolmogorov 1941)
interpolator in the Geostatistical Analyst extension of ArcGIS. The
interpolation was based on the original climate databases of the
public domain Worldclime supported by the University of Berkley
(http://www.biogeo.berkeley.edu/)
for the period 1950-2000 (Hijmans et al. 2004) and the DEM.
Model building
Building ecological
niche models to predict species ranges has been one of the main
focuses over the last 10 years in landscape ecology and conservation
biology (Peterson & Cohoon 1999). Special concern has been
devoted to develop species ranges in poorly known regions where only
presence points are available. Several analytical approaches have
been applied to these challenges, varying from simple sets of rules
based on overlays of environmental and species occurrences data
creating a so-called "environmental envelop" (Krabbe et
al. 1998) to sophisticated multivariate analyses such as
mahalanobis distance (Cuesta et al. 2003) or logistic
regression (Loiselle et al. 2003).
Among these methods,
the Genetic Algorithm for Rule-Set Prediction (GARP, Stockwell &
Noble 1992; Stockwell & Peters 1999) is being increasingly used
in predictive models of species distributions. GARP is an artificial
intelligence-based super-algorithm that works by combining sets of
rules to build the most accurate prediction possible for the region
being considered. GARP has been used, among other applications, to
test different biogeographic theories (Anderson et al. 2002b),
to evaluate colonization processes by invasive species and parasites
(Peterson & Vieglais 2001; Peterson & Shaw 2003), and to
assess the effects of climate change in the distribution of species
(Peterson et al. 2001).
SDMs were developed
using a PC desktop-implementation of the program GARP
(http://www.lifemapper.org/desktopgarp/)
following the optimization parameters suggested by Anderson et al.
(2003), Loiselle et al. (2003), and Raxworthy et al.
(2003). For species with more than 20 unique occurrence records, the
records were randomly divided in two subsets: training points (75% of
the species records) and testing points (25% of the points). For
species with less than 20 occurrences, all the points were used as
training points to develop the models.
In all cases, an
algorithm convergence threshold of 0.01 was selected with 1,000
iterations as an upper limit for each run. Each model was set to
perform 100 tasks in order to account for the elements of
stochasticity inherent to genetic algorithms, which means GARP
produces slightly different models in every run (Stockwell and
Peterson 2002; Peralvo 2004).
Species distribution
models were obtained based on the selection of the 10 best-fit models
(the 10 runs best subset). The best subsets for each species were
selected by means of Anderson et al. (2003) criteria for
selecting optimal distribution models, which states that model
performance follows an inverse relationship between omission and
commission errors and that best models in general shows low omission
errors and moderate commission indexes.
Thus, an omission
threshold of 5% and a commission threshold of 50% of the distribution
were set for each model. For species with less than 20 occurrence
points, intrinsic measures of omission and commission were selected,
whereas extrinsic measures were used for species with more than 20
occurrence points. Afterward, the 10 models included in the best
subset were summed to create a composite map, and the final modeled
range for each species was generated using the pixels that were
predicted as part of the distribution in the 10 models. These models
are referred to as the "potential distributions" of each
species hereafter (Peralvo 2004).
SDMs evaluation
Model accuracy was
evaluated based on a sensitivity analysis using extrinsic and
intrinsic measures (Cumming 2000). Areas of predicted absence where
the species has been recorded are regarded as omission errors (Type
II error) or "false absence". Areas predicted as presence
by the model but where the species has not been observed constitute
commission errors (Type I error) or "false presence".
False absence value is particularly useful because it measures the
amount of unexplained variation in the data (number of residuals);
the greater the values of false negatives, the more models are likely
to be unrealistic.
Based on this
criterion, the mean values of intrinsic omission error and extrinsic
commission index were evaluated for the 10 models in the best subset
together with the intrinsic overall accuracy and the percentage of
non predicted area within the study area (Table 3). The intrinsic
overall accuracy is a descriptive statistic derived from the
confusion (error) matrix; where "a" and "d"
represents the areas (number of pixels) correctly predicted as
presence or absence. Conversely, "c" and "d"
represents the omission and commission errors. This quantity ranges
from 0 to 1.
Finally, for those
target species for which a test dataset was available (n ≥ 20) a
one tiled chi-squared test was performed. The significance of each
model in the best subset was tested using a null hypothesis that no
relationship existed between the test dataset and the predicted
distributions.
Results

Species distribution models (SDMs)
We developed 138
SDMs (112 plant and 26 bird species) based on 1933 unique occurrence
records for plant species and 517 points for bird species. The
minimum number of records for a plant species was four (median = 11)
and five (median = 18) for birds; the maximum number reached 80
unique records for a plant species (Cavendishia bracteata) and
54 for a bird (Grallaria rufula rufula). 71% and 54% of
the modeled plant and bird species had less than 20 independent
records.
Measures of model performance
a. Composite
measures of performance
For those models
with 20 or more points extrinsic performance measures (χ2)
were almost always significant. All thirteen models for birds showed
significant deviations from random predictions (χ2 =
7.31 - 71.52; p << 0.05). Thirty of 33 models for plants
were highly significant (χ2 = 2.79 - 64.71; p <<
0.05); the other 3 models had non-significant departure from the
desired direction.
When comparing both
plant and bird models with 20 or more points, a consistent trend was
observed between intrinsic and extrinsic measures of overall model
performance. The correlation analysis shows a highly significant
relationship of both composite measures (r = 0.83; p < 0.01). The
regression analysis explained the variation among both measures in
70% of the cases (Figure 2) showing a consistency on both indicators of
model performance. Yet, variation in overall internal accuracy was
minimal compared with a great variation in the extrinsic accuracy
parameter. This may be related by the high susceptibility of the
chi-square statistic towards the amount of test points and to the
proportional extent of predicted presence. Thus, bird models in
general had relative large number of test points which resulted in a
high statistical power (Figure 3).
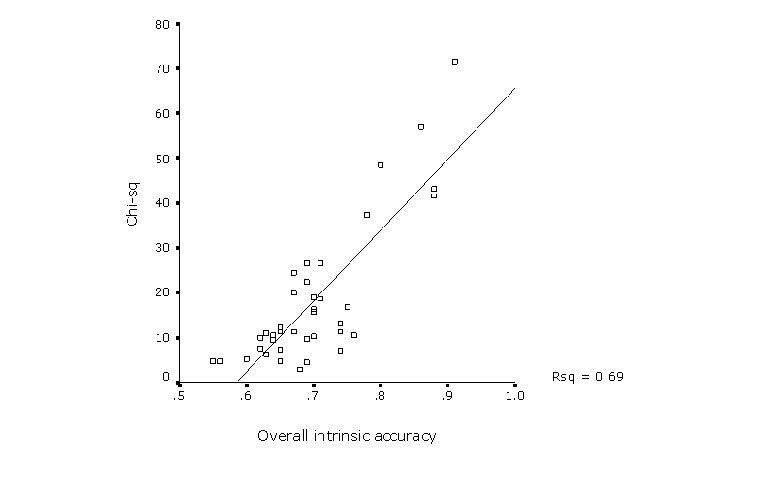
Figure 2. Regression analysis to test for a consistency between intrinsic and extrinsic measures of overall model performance
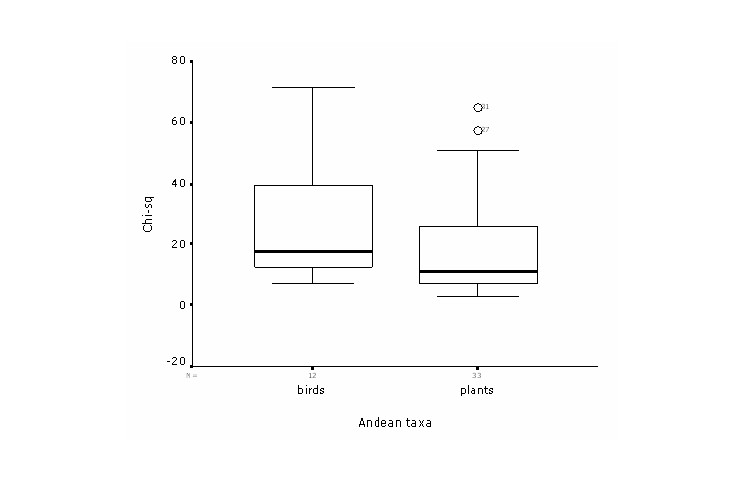
Figure 3. Boxplot
comparing chi-square statistical power among birds and plant
models.
b. Intrinsic
omission and commission errors
Within intrinsic
values for omission index of the plants models did not track their
corresponding commission values. Still, a modest negative correlation
exists among these two variables (r = -0.58; p ( 0.01). The
highest percent (93%) of the cases are clustered at omission values <
0.1, while the dispersion of the values within the commission axis is
substantial with a great concentration of cases on the range between
0.75 and 1.0 (median = 0.79). A peculiar relationship is observed at
the lower right side of the scatterplot, where all the cases with a
commission value of over 0.70 had no training localities falling
outside the predict area ((Figure 4a)).
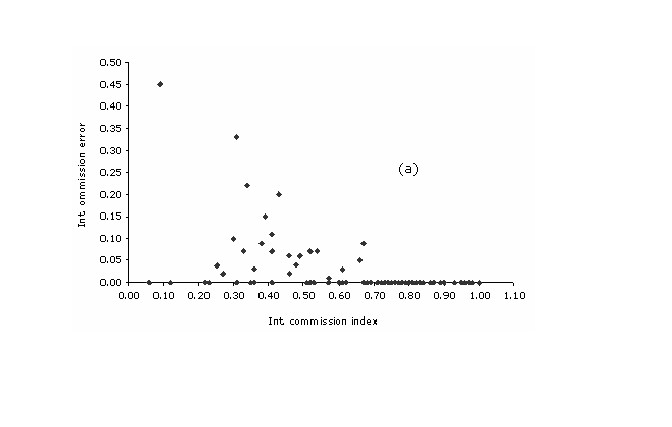
Figure 4a. Intrinsic plots of omission error vs. commission index, for each of the (a) plant and (b) bird species.
This pattern might
be explained by the strong relationship of the non-predicted area of
the models with the internal commission index. Correlations between
the two measures for the plant models were high (r = 0.89; p <
0.01), with highly significant regression (Rsq = 0.78; p < 0.001).
All the cases with a commission index > 0.7 and a omission error
of 0.0 have less than 20 occurrence points and, in the majority of
the cases, had small geographic ranges (< 10.000 km2)
and a mean overall accuracy of 0.51.
0.75) and the lowest omission error
values (((Figure 4b)).
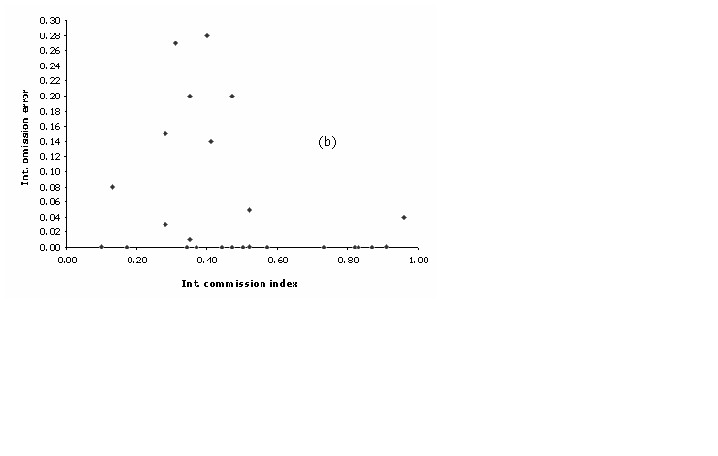
Figure 4b. Commission Index.
Correspondingly to
the plant models, the latter pattern can be explained by contrasting
the commission index with the percentage of area non-predicted of
each model. A discrete positive correlation was observed among these
two variables (r = 0.64; p < 0.01) suggesting that high commission
values were partially related to a poor capacity of GARP on
describing the realized niche of that particularly species. This
analysis highlights the fact of 5 specific models that might have
problems due to its high level of uncertainty. All these models have
less than 20 training points and all but one (Eriocnemis luciani)
present a restricted geographic extent (< 10,000 km2).
Discussion

Species ranges
Measures
of model performance
Our results
demonstrate the ability of GARP models to predict Andean species
distributions over large geographic extents. GARP models are
applicable to a variety of species with different ecological
requirements and under different sample scenarios. Furthermore,
predictions were tested in areas outside the spatial extent of the
training data with high accuracy. Thus, ecological niche modelling is
a promising tool for understanding species distributions in poorly
known regions.
Our
tests showed that internal overall accuracy measures help to evaluate
the overall model performance. Yet, it fails as a measure of quality
to discriminate among a suite of final GARP models. Although we
demonstrated that GARP models can be used under different scenarios,
the exploration of the model quality is rather complex, due to the
high dispersion of the statistical measures between different
scenarios. Models with more than 20 records showed a high dispersion
of the chi-square statistic which makes it difficult to establish
effective means for identifying the best models. According to
Peterson et al. (2001) extrinsic model significance (χ2)
probably varied among species in part due to the power afforded when
varying sample sizes in the test data sets, and because of the
relative extent of suitable habitat for each species. Conversely, in
our study, the internal overall accuracy varied little among models.
Stockwell & Peterson (2002b) and Anderson et al. (2003)
also found that overall accuracy reached an apparent plateau with
sample sizes of 20-50 localities. Thus, this measure helps to
evaluate overall model performance but fails as a measure of quality
to discriminate among a suite of final GARP models.
Likewise,
the use of omission and commission errors may be useful for assessing
overall model performance and to discriminate the best ones among
several GARP outputs, because of the relative high sensitivity of the
overall accuracy tests to sample sizes (Anderson et al. 2003).
This quantities allowed us to evaluate models developed with no test
points because patterns in intrinsic measures are repeated in the
independent extrinsic ones; hence intrinsic measures hold potential
for assessing model quality when all available data points are used
for model construction.
The
majority of the models for the widespread species combined low
measures of omission with mid to fairly high levels of commission,
and a fairly high overall intrinsic accuracy. These results concur
with the findings of Peterson et al. (2002) and Anderson et
al. (2003) who stated that in the best GARP models large areas
are included as predicted presence (yielding high values in the
commission index) in order to reduce omission to acceptable levels
without overfitting the data.
The majority of the
models for restricted range species showed extreme low omission
values, extreme high commission values, and a relative mid overall
intrinsic accuracy. The high commission values together with high
number of cells where the algorithm could not define whether the
species is present or absent, suggest a high degree of uncertainty in
the produced models.
Consequently,
the produced models overfitted the data by including most or all of
the training points and small regions around them which mean GARP
models included the core ecological distribution of that species, but
excluded ecologically peripheral parts of possible distribution.
According to Anderson et al. (2003) for these species (p.e.,
Grallaria ridgleyi; Ceratostema oellgardii), the best
GARP models should minimize omission without increasing commission
excessively (because pixels of predicted presence represent a small
fraction of the study region) which was not the case for 35% of
produced models.
Conclusions

a. Ecological modeling methods
Our results suggest
that ecological niche modeling methods to predict the geographic
distribution of Andean-centered taxa constitute a promising tool
which allow evolutionary, biogeographic and conservation research
based on data from specimens present in natural history collections.
Predictive models of species distributions take advantage of the fact
that museum and herbaria collections tend to be more biased in
geographic space than in environmental space. Thus, predictive models
allow estimating species' presence/absence patterns for large
geographic areas, optimizing the use of existing presence-only
datasets. The modeling system used in this study proved to be
especially well suited for this application because in the majority
of the cases its predictions were not affected by the spatial biases
present in the biological databases.
Another advantage of
using species distribution models as predictors of biodiversity
patterns within the study area is the improvement in spatial
resolution in relation to higher order surrogates such as vegetation
types. Vegetation types assume a homogeneous distribution of an
assemblage of species in each class, whereas species richness based
on modeled distributions is likely to capture the heterogeneous
nature of environmental variation more faithfully (Peralvo 2004). In
this sense, predictive models work better than highly generalized and
subjectively constructed estimations of species distributions, thus
provide a systematic framework to evaluate distribution of species
that can be quantitatively tested using independent data. Finally the
selection of ecological meaningful taxa over a significant
biogeographic unit allowed us to delineated, with a high accuracy
level, areas of high endemism on the Eastern Cordillera.
The main limitations
associated with predictive models are related to (1) the scarce
quantity of independent species records, (2) the coarseness of the
environmental predictors, and (3) the static nature of the modeling
techniques derived from the necessary assumption of equilibrium.
First, model accuracy tends to increase with sample size (e.g.
Peterson et al. 2001; Stockwell & Peterson 2002a);
however, the minimum sample size relays on the modeling method
applied, the biology of the species, the environmental predictors
used, and the environmental characteristics of the study area
(Peterson et al. 2002). Nevertheless, setting an objectively
threshold sample size requires testing the increment in predictive
accuracy achieved at different sample sizes until a plateau in
accuracy is reached (Anderson et al. 2002b; Stockwell & Peterson
2002a). Yet, this procedure requires relatively large sample sizes to
test and train the models which are not the case for most rare and
endemic species of the Andes. Consequently most studies that use
biological collections as primary data sources have followed a rather
arbitrary approach to the definition of minimum sample size
requirements as in our case.
Second, the climatic
variables used in this study are rather coarse, thus they might
obscure specific environmental gradients which in turn are function
of the species' fundamental niche. Finally, correlative
modeling approaches do not allow the inclusion of dynamic processes
that have important consequence in the distribution of species. As a
result, models present commission and omission errors that reflect
the uncertainty introduced by factors such as environmental and
demographic stochasticity. In spite of these limitations, correlative
models are appropriate when a preliminary assessment of the
distribution patterns of species is required over large geographic
areas, since at these scales bioclimatic factors play a fundamental
role as controls of species distributions.
Acknowledgements

The reviews of R.
Hofstede, J. Duivenvoorden, and S. Benítez helped greatly to
improve the manuscript and are appreciated. We thank Frank van Manen,
Laura Thomson, and Angela Parrado for their comments and language
editing on early version of the manuscript. Our special gratitude to
Juan Fernando Freile, Lorena Endara, James Luteyn, Kerem Boyla,
Catherine Graham, Julio Betancourt, and Isidoro Sánchez for
providing information and sharing their knowledge on the distribution
of the selected taxa within the Eastern Cordillera. We would like to
acknowledge all the institutions that made possible this research:
The Nature Conservancy-Ecuador, EcoCiencia, Fundación Natura,
Conservation International-Ecuador, Instituto Alexander von Humboldt,
World Wildlife Fund-Colombia, Instituto Nacional de
Ciencias-Universidad Nacional de Colombia, Birdlife International,
Universidad de Cajamarca, Herbario Nacional (QCN-E), Corporación
Ornitológica Ecuatoriana (CECIA), Chicago Field Museum of
Natural History (FMNH), Academy of Natural Sciences of Philadelphia
(ANSP), the Berkeley Museum of Natural History, and all the
institutions that continuously provide information to the Tropicos
Database stored at the Missouri Botanical Garden (MOBOT) web-browser
search engine. This research was supported by The Nature
Conservancy-Ecuador and the MacArthur foundation.
References

Anderson, R P., M.
Laverde & A.T. Peterson. 2002a. Geographical distributions
of spiny pocket mice in South America: Insights from predictive
models. Global Ecology and Biogeography 11: 131-141.
Anderson, R.P., M.
Laverde & A.T. Peterson. 2002b. Using niche-based GIS modeling to
test geographic predictions of competitive exclusion and competitive
release in South American pocket mice. Oikos 98: 3-16.
Anderson, R.P., D.
Lew & A.T. Peterson. 2003. Evaluating predictive models of
species' distributions: Criteria for selecting optimal models.
Ecological Modeling 162: 211-232.
Anderson, R.P. &
E. Martinez-Meyer. 2004. Modeling Species' distributions for
preliminary conservation assessments: an implementation with the
spiny pocket mice (Heteromys) of Ecuador. Biological Conservation
116: 167-179.
Avise, J.C. 2000.
Phylogeography: the history and formation of species. Harvard
University Press, Cambridge, Mass. USA.
Bleiweiss, R. 1998a.
Tempo and mode of hummingbird evolution. Biological Journal of the
Linnean Society 65: 63-76.
Bleiweiss, R. 1998b.
Origin of hummingbird faunas. Biological Journal of the Linnean
Society 65: 77-97.
Boone, R.B. &
W.B. Krohn. 2000. Partitioning sources of variation in vertebrate
species richness. Journal of Biogeography 27: 457-470.
Borchsenius, F.
1997. Patterns of plant species endemism in Ecuador. Biodiversity
and Conservation 6: 379-399.
Croat T.B. 1991. A
revision of Anthurium section Pachyneurium (Araceae). Annals
of the Missouri Botanical Garden 78: 539-855.
Cuesta, F., M.
Peralvo & F. van Manen. 2003. Andean bear habitat use in the
Oyacachi River Basin, Ecuador. Ursus: 14: 198-209.
Cumming, G.S. 2000.
Using between-model comparisons to fine-tune linear models of species
ranges. Journal of Biogeography 27: 441-455.
Dinerstein,
E., D.M. Olson, D.J. Graham, A.L. Webster, S.A. Primm, M.P.
Bookbinder & G. Ledec. 1995. A conservation assessment of the
terrestrial ecosystems of Latin America and the Caribbean. World
Wildlife Fund and World Bank, Washington, DC.
Duellman,
W. & E. Wild. 1993. Anuran amphibians from the Cordillera
Huancabamba, Northern Peru: Systematics, ecology, and biogeography.
Occasional papers of the Museum of Natural History. The
University of Kansas, Lawrence 57: 1-53.
Erwing,
T. 1991. An evolutionary basis for conservation strategies. Science
253: 750-752.
Fjeldså,
J. 1995. Geographical patterns of neoendemic and older relict species
of Andean forest birds: the significance of ecologically stable
areas. in Churchill S.P., H. Balslev, E. Forero & J. L. Luteyn
(Eds). Biodiversity and conservation of Neotropical Montane
Forests - proceedings of the Neotropical Montane Forest
Biodiversity and Conservation Symposium, the NYB, 21-26
June.1993. The Ney York botanical Garden. NY. Pp: 1-667.
Fjeldså,
J. & N. Krabbe. 1990. Birds of the high Andes - A manual
to the birds of the temperate zone of the Andes and Patagonia, South
America. Zoological Museum - University of Copenhagen and
Apollo Books. Svendborg.
Fjeldså,
J., E. Lambin & B. Mertens. 1999. Correlation between endemism
and local ecoclimatic stability documented by comparing Andean bird
distributions and remotely sensed land surface data. I 22:
63-78.
García-Moreno,
J., P. Arctander & J. Fjeldså. 1999. Strong Diversification
at the treeline among Metallura hummingbirds. The Auk 116:
702-711.
Gentry,
A. 1982. Neotropical Floristic Diversity: Phytogeographical
connections between central and south america, pleistocene climatic
fluctuations, or and accident of the Andean orogeny. Annals of the
Missouri Botanical Garden 69: 557-593.
Guisan, A. 2002.
Semiquantitative response models for predicting the spatial
distribution of plant species. in Scott, J.M., P.J. Heglund &
M.L. Morrison (Eds). Predicting species occurrences: issues of
scale and accuracy. Pages 315-326 Island Press, Washington, D.C.
Guisan,
A. & N.E. Zimmermann. 2000. Predictive habitat distribution
models in ecology. Ecological Modeling 135:147-186.
Heindl,
M. & K.L. Schuchmann. 1998. Biogeography, geographical variation
and taxonomy of the Andean genus Metallura, Gould, 1847.
Journal für Ornithologie 139:
425-473.
Hijmans,
R.J., S. Cameron & J. Parra. 2004. WorldClim,
version 1.2. A square kilometer resolution database of global
terrestrial surface climate. Available at
http://biogeo.berkeley.edu/
Http://www.biogeo.berkeley.edu/
Http://www.lifemapper.org/desktopgarp/
Http://www.nybg.org/bsci/res/lut2/
Http://seamless.usgs.gov/
Huston,
M.A. 2002. Introductory Essay: Critical Issues for Improving
Predictions. in Scott J.M., P.J. Heglund & M.L. Morrison (Eds).
Predicting species occurrences: issues of scale and accuracy.
Pages 7-21 Island Press, Washington, D.C.
Hutchinson,
G.E. 1957. Concluding remarks. Cold Spring Harbor Symposium in
Quantitative Biology 22: 415-427.
Jorgensen,
P.M., D. Neill, & S. León-Yánez. 1999.
Introduction. in Jorgensen P.M. & S. León-Yánez.
(Eds). Catalogue of the vascular plants of Ecuador. Herbarium
de la Universidad Católica del Ecuador, Herbarium of the Museo
Ecuatoriano de Ciencias Naturales, and Department of Systematic
Botany of Aahrus University. Pages 1-108 Missouri Botanical
Garden Press, St. Louis.
Jorgensen,
P.M. & S. Leon Yanez. (Eds). 1999. Catalogue of the vascular
plants of Ecuador. Missouri Botanical Garden Press, St. Louis.
Jorgensen, P.M., C.
Ulloa-Ulloa, J. Madsen &R. Valencia. 1995. A floristic analysis
of the high Andes of Ecuador. in S.P. Churchill, H. Balslev, E.
Forero & L. Luteyn (Eds). I. Pages 221-237 The New York
Botanical Garden, New York, USA.
Kolmogorov,
A.N. 1941. Interpolation and extrapolation of stationary random
sequences. Isvestiia Akademii Nauk SSSR, Seriia Matematicheskiia
5: 3-14.
Krabbe,
N., F. Skov, J. Fjeldså &I .K. Petersen. 1998. Avian
diversity in the Ecuadorian Andes. Centre for Research on
Cultural and Biological Diversity of Andean Rainforest (DIVA), 143
pp. DIVA Technical Report n°4. Denmark.
Leimbeck,
R.M., R. Valencia & H. Balslev. 2004. Landscape diversity
patterns and endemism of Araceae in Ecuador. Biodiversity and
Conservation 13: 1755-1779.
Loiselle,
B.A., C.A. Howell, C.H. Graham, J.M. Goerck, T. Brooks, K.G. Smith &
P.H. Williams. 2003. Avoiding pitfalls of using species distribution
models in conservation planning. Conservation Biology 17:
1591-1600.
Luteyn,
J. 2002. Diversity, adaptation and endemism in neotropical Ericaceae:
biogeographical patterns in the Vaccinieae. The Botanical Review
68: 55-87.
Missouri Botanical
Garden (MBG). 2004. MOBOT - W3 TROPICOS.
Myers,
N., R.A. Mittermeier, C.G. Mittermeier, G.A.B. da Fonseca & J.
Kent. 2000. Biodiversity hotspots for conservation priorities. Nature
403: 853-858.
Peralvo, M.F. 2004.
Identification of biodiversity conservation priorities using
predictive modeling: An application for the tropical dry forests of
western Ecuador and northern Peru. M.Sc. Thesis. University of
Texas. Austin.
Peterson, A.T.,
Ball, L.G. & K.P. Cohoon. 2002. Predicting distributions of
Mexican birds using ecological niche modeling methods. Ibis
144: 27-32.
Peterson, A.T. &
P. Cohoon. 1999. Sensitivity of distributional prediction algorithms
to geographic data completeness - Short communication -.
Ecological Modelling 117: 159-164.
Peterson,
A.T., V. Sanchez-Cordero, J. Soberon, J. Bartley, R.H. Buddemeier &
A.G. Navarro-Siguenza. 2001. Effects of global climate change
on geographic distributions of Mexican Cracidae. Ecological
Modeling 144: 21-30.
Peterson, A.T. &
J. Shaw. 2003. Lutzomyia vectors for cutaneous leishmaniasis
in Southern Brazil: ecological niche models, predicted geographic
distributions, and climate change effects. International Journal
for Parasitology 33: 919-931.
Peterson, A.T. &
D.A. Vieglais. 2001. Predicting species invasions using ecological
niche modeling. BioScience 51: 363-371.
Raxworthy, C.J., E.
Martinez-Meyer, N. Horning, R.A. Nussbaum, G.E. Schneider, M.A.
Ortega-Huerta & A. Townsend-Peterson. 2003. Predicting
distributions of known and unknown reptile species in Madagascar.
Nature 426: 837-841.
Rice, N. 2005.
Phylogenetic relationships of Antpitta genera (Passeriformes:
Formicariidae). The Auk 122:673-683.
Ridgley, R. & P.
Greenfield. 2001. The birds of Ecuador with the collaboration
of M. Robbins and P. Coopmans. Cornell University Press. Ithaca, NY.
Schuchmann, K., A.
Weller & I. Heynen. 2001. Systematics and biogeography of the
Andean genus Eriocnemis (Aves: Trochilidae). Journal fuer
Ornithologie 142: 433-481.
Stockwell, D.R.B. &
A. T. Peterson. 2002a. Effects of sample size on accuracy of species
distribution models. Ecological Modeling 148: 1-13.
Stockwell, D.R.B. &
A. T. Peterson. 2002b. Controlling bias in biodiversity data. in
Scott, J. M., P.J. Heglund & M.L. Morrison et al. (Eds).
Predicting species occurrences: issues of scale and accuracy.
Pages 537-545 Island Press, Washington D.C.
Stockwell, D.R.B. &
D.P. Peters. 1999. The GARP modeling system: Problems and solutions
to automated spatial prediction. International Journal of
Geographic Information Systems 13: 143-158.
Stockwell, D.R.B. &
I.R. Noble. 1992. Induction of sets of rules from animal distribution
data: A robust and informative method of analysis. Mathematics and
Computers in Simulation 33: 385-390.
Vargas, J.H., T.
Consiglio, P.M. Jorgensen & T.B. Croat. 2004. Modelling
distribution patterns in a species-rich plant genus, Anthurium
(Araceae), in Ecuador. Diversity and Distribution 10: 211-216.
Williams, P.H., C.
R. Margules & D.W. Hilbert. 2002. Data requirements and data
sources for biodiversity priority area selection. Journal
of Biosciences 27: 327-338.
World
Wildlife Fund (WWF). 2001. Visión de la biodiversidad de
los Andes del Norte. Santiago de Cali, Colombia (In
Spanish).
Young, K., C. Ulloa
Ulloa, J. Luteyn & S. Knapp. 2002. Plant Evolution and Endemism
in Andean South America: An Introduction. The Botanical Review
68: 4-21.
Appendices:
Tables
Table
1. Andean-centered taxa used as descriptors of the alpha diversity
patterns on the Eastern Andean ridge (Table (a) refers to vascular
plants and, table (b) to bird taxa).
(a)
|
Andean
genera
|
Species
recorded in Ecuador
|
Species
recorded in Eastern cordillera
|
Percentage
(%)
|
Endemic
species recorded in Eastern cordillera
|
Percentage
(%)
|
|
Anthurium
|
227
|
55
|
24.2
|
11
|
20.00
|
|
Lepanthes
|
287
|
194
|
67.6
|
118
|
60.82
|
|
Cavendishia
|
21
|
13
|
61.9
|
2
|
15.38
|
|
Ceratostema
|
28
|
28
|
100.0
|
26
|
92.86
|
|
Psammisia
|
26
|
17
|
65.4
|
5
|
29.41
|
(b)
|
Andean
genera
|
Species
recorded in Ecuador
|
Species
recorded in CRO
|
Percentaje
(%)
|
Endemic
species recorded in CRO
|
Percentage
(%)
|
|
Grallaria
|
15
|
12
|
80.00
|
2
|
16.67
|
|
Eriocnemis
|
7
|
5
|
71.43
|
2
|
40.00
|
|
Metallura
|
6
|
4
|
66.67
|
1
|
25.00
|
|
Coeligena
|
7
|
4
|
57.14
|
0
|
0.00
|
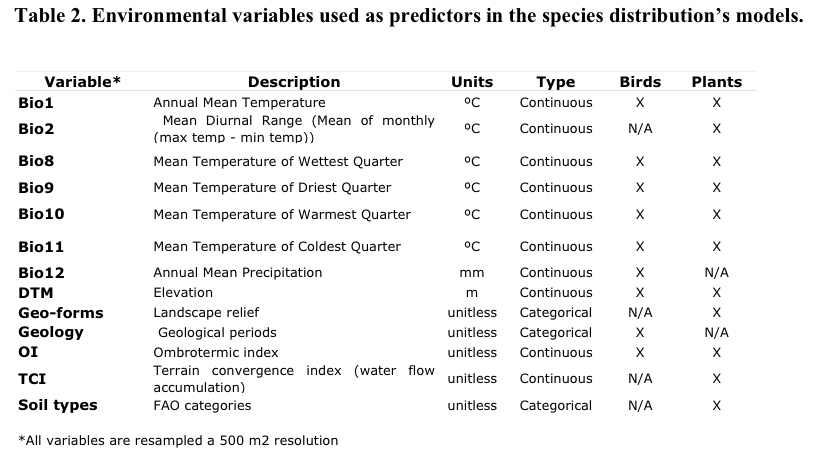
Table 2. Environmental variables used as predictors in the species distribution's models.

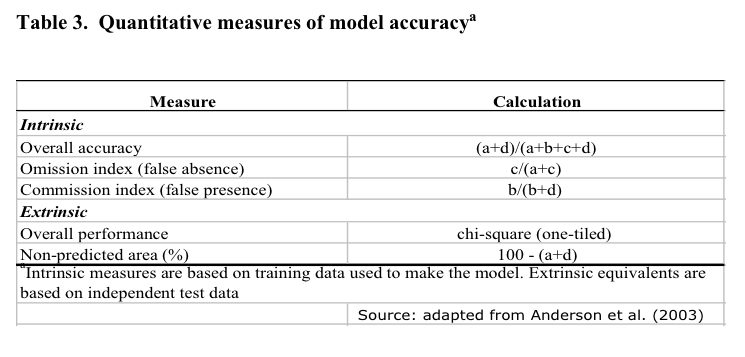
Table 3. Quantitative measures of model accuracy

-21-



